Variant Interpretation Pipeline (VIP)¶
VIP is a flexible human variant interpretation pipeline for rare disease using state-of-the-art pathogenicity prediction (CAPICE) and template-based interactive reporting to facilitate decision support.
The VIP pipeline can be used starting from either your fastq, bam/cram or .g.vcf/vcf data,
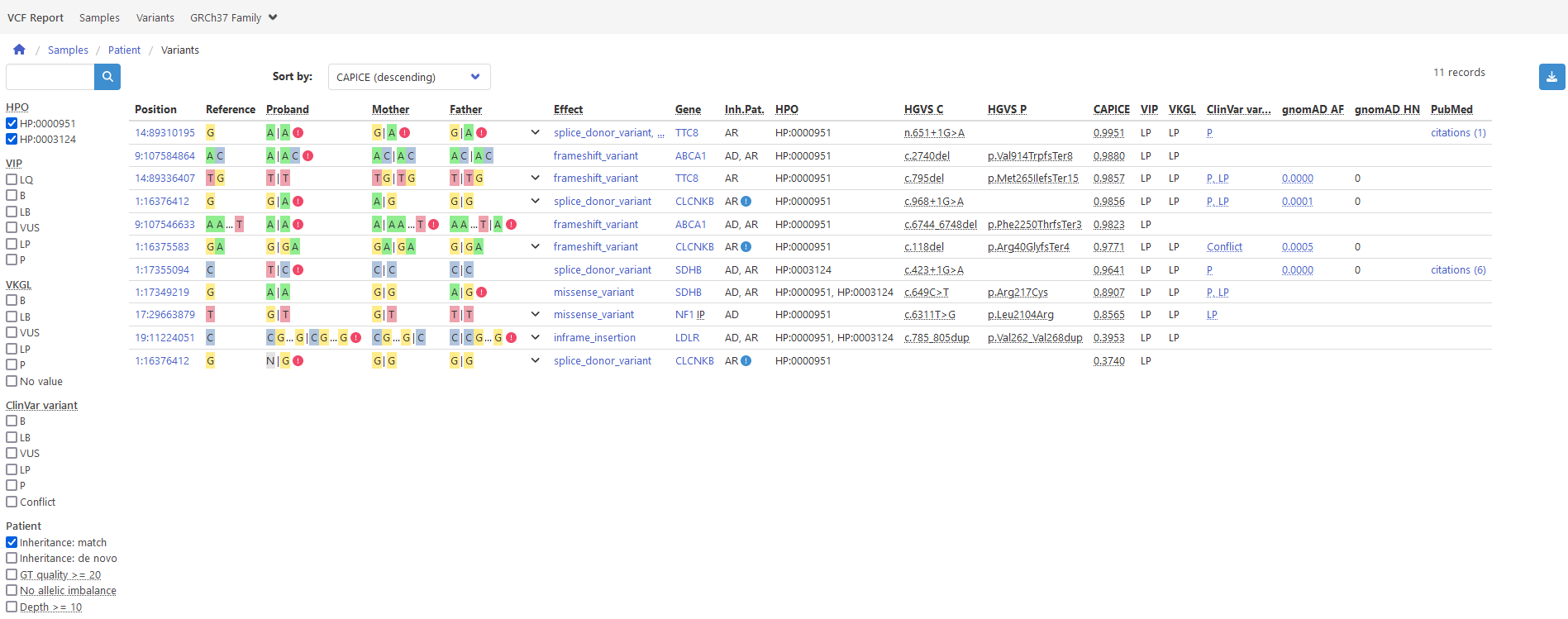
every entry point will result in a vcf file with your annotated, classified and filtered variants
as well as a interactive HTML report with the same variants, prioritized by the CAPICE pathogenicity score
and providing additional aids like a genome browser and a representation of the decisions leading to the VIP
classification.
VIP can be used for single patients, families or cohort data.
Try it yourself
Visit https://vip.molgeniscloud.org/ to analyse your own variants
Tip
Article published in NAR Genomics and Bioinformatics
 ]
]
Above: report example
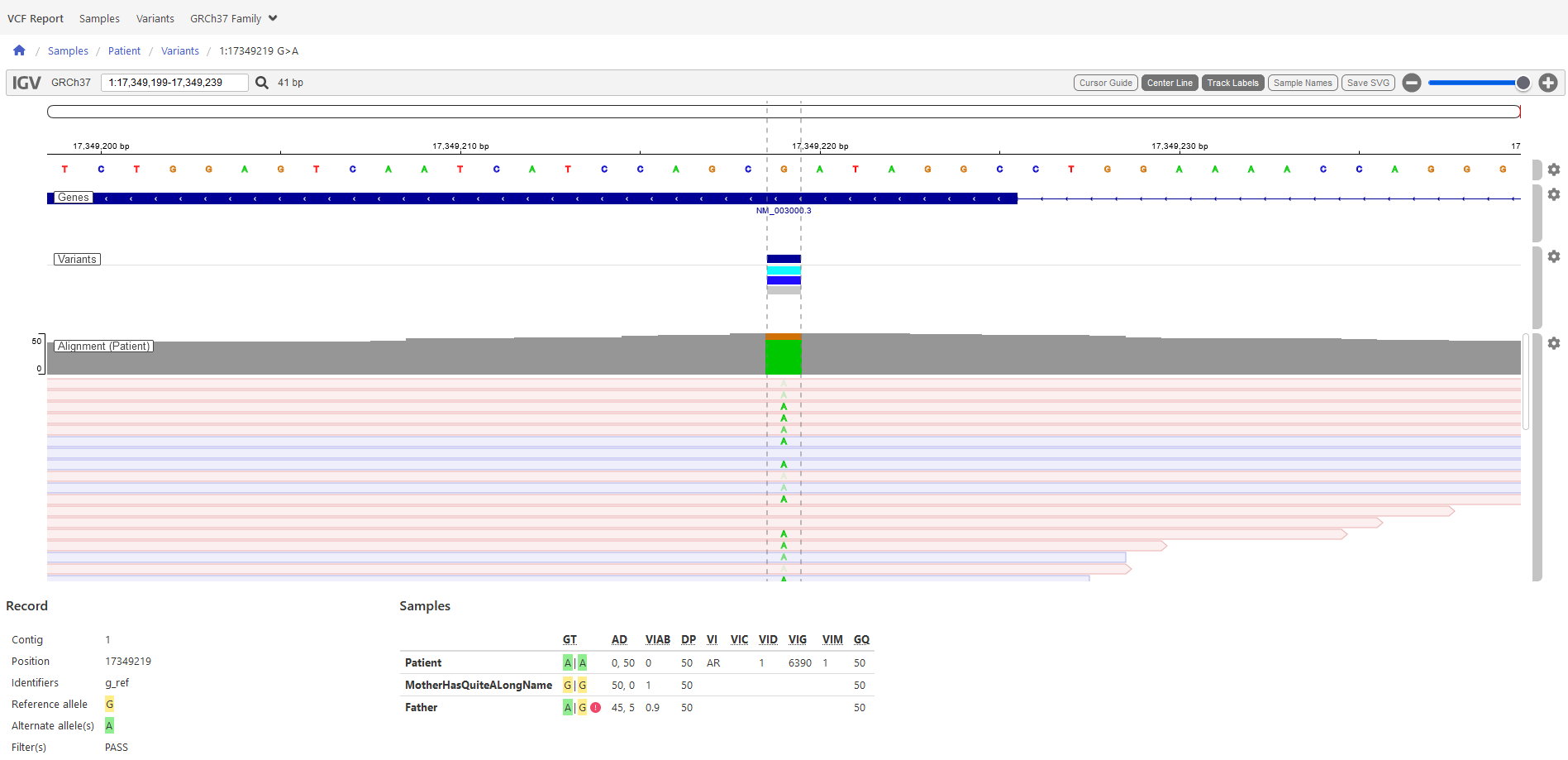
Above: report example: genome browser